Humans, bacteria, and viruses all use a common genetic code to communicate with one another, but new evidence shows that some viruses found in the human gut may use an alternative genetic code to encode their proteins.
Credit: Philip Patenall/Macmillan Publishers Limited
There are over 7,000 languages in use in the world today, and many people speak more than one of these languages. From Arabic to Zulu, it’s not strange for us to think that there are multiple languages used to describe the world around us and to transfer information from one generation to another. However, in the genetic world, there’s one standard language that all organisms understand: the genetic code. This is the language that almost all organisms use to read RNAs and turn them into proteins. Yet, just like we have many languages, there are also multiple genetic codes, but when and where they’re used is still an area of study.
These genetic languages are more like a code or cipher than the written and spoken languages we typically think about. Each “word” of this language, called a codon, is represented by three letters. In total there are 21 possible words, each of which represents an amino acid, the building blocks of proteins. Any given string of words, akin to a sentence, is contained within an RNA molecule, and when a ribosome reads a sentence in a process called translation, it can create the encoded protein.
Because all organisms use the standard genetic language or code to create proteins from RNA, this allows bacteria and viruses to infect our cells and use our cellular machinery to make their own proteins. The ribosomes in our cells are able to read the RNA of these pathogens because they’re written in a language that our cells understand. This overlap doesn’t just apply to pathogens that infect humans, it also comes into play when viruses infect bacterial cells. While seemingly unimportant to humans, these viruses can play a role in human health by modulating the gut microbiome, and they’ve even been used to treat antibiotic resistant bacterial infections.
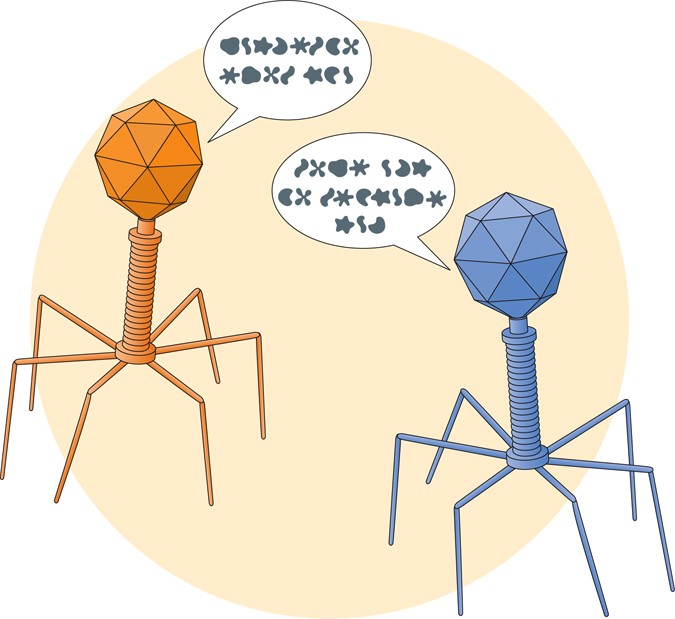
Viruses that infect bacteria are called bacteriophages, and these are the viruses we think of when we try to picture a virus – a hexagonal head on top of a pair of legs. These bacteriophages use the same genetic code as bacteria, allowing their viral proteins to be created within a bacterial host. The consequences of this are similar to when a virus infects a human; the production of viral proteins ultimately leads to immune system activation and subsequent killing of the bacterial cell.
Recently, however, studies have found that these viruses may use one of the alternative languages, or genetic codes, to encode some of their proteins. This means that the three letter word in the standard genetic code which means red would be translated as jacket in this alternative genetic code. In a recent Nature Communications study by Peters and colleagues, researchers used a combination of RNA and protein sequencing to uncover evidence that bacteriophages in the human gut use an alternative language to encode information.
Three of the words in the genetic code are used to indicate the end of the sentence, similar to the words “the end” or “fin” in the English language. These words are aptly called stop codons. In the case of protein translation, the stop codon stops the ribosome from reading further on the RNA molecule and allows the protein to be released from the ribosome. Typically, the codon TAG encodes this stop codon. However, the bacteriophages studied in this paper can change the interpretation of this TAG codon from a stop into the amino acid glutamine. This means that the sentence won’t end where expected, and it will instead make a longer sentence that has a different meaning than the original.
The authors of this paper were able to discover the use of this alternative translation by comparing the RNA sequence of bacteriophages to the protein output. Like translating one language into another, the authors looked at the sequence of the RNA molecule and used the known genetic code to determine what the amino acid sequence generated from that RNA molecule should be. If the known genetic code was used to produce the proteins, then these amino acids should be found when protein sequencing is performed. However, the authors found that these predicted amino acids were not showing up in their sequencing. Instead, different amino acids that were not predicted by the genetic code were appearing especially at what were formerly known as TAG stop codons. The only way to explain this was through the use of an alternative genetic code; a unique language. When the authors used alternative genetic codes to translate the RNA molecule into protein, they found that only one genetic code was able to predict the protein output: genetic code 15. Interestingly, the bacteriophages only used this alternative language later on in their life cycle, likely to temporally control the production of specific viral proteins.
This important discovery now provides us with evidence that bacteriophages in the human gut can use different languages to encode their proteins. While there are 33 known variations of the genetic code used by a plethora of organisms, this study is the first evidence of bacteriophages using genetic code 15. These alternative genetic codes provide us with another tool in which to understand bacteriophage life cycles. Previously, scientists were trying to translate virus genomes into protein by only using the standard genetic code, but as this study shows, viruses can use alternative codes to make their proteins. This is akin to trying to translate a Spanish text into English using a dictionary with the pages missing – while some of the sentences can be translated perfectly, others will have errors or completely untranslatable words. By discovering the use of genetic code 15 by viruses, we’re now able to better interpret what’s written in the genetic book of viruses. This work ultimately expands our understanding of viruses and can provide insight into how we might design therapies in the future to address both viral and antibiotic resistant infections.
Edited by Sarah Lester and Jayati Sharma



Leave a comment